INTRODUCTION
Tissue engineering aims for assembling functional constructs that restore, maintain, or improve damaged tissues or whole organs. In the quest for the optimal tissue-engineered solution per specific application, the employment of gene expression analysis techniques is commonly desired. A pre-requisite for gene expression analysis is the isolation of high purity RNA. Methods and protocols for the isolation of highly pure RNA from cells and tissues are well established, and ready-to-use reagents are available from a number of commercial suppliers. However, these reagents and corresponding protocols perform poorly when sample composition differs from the standards of biological tissues and organs. The high quantities and diverse nature of biomaterials typically contained in tissue-engineered constructs usually challenge the performance of well established RNA isolation reagents and protocols, making optimisation a must per specific application. Hybrid thermoplastic-hydrogel cell-laden constructs manufactured by means of 3D bioprinting are becoming a popular choice for tissue engineering of a wide range of tissues; specially those subject to mechanical load. Herein, a protocol for isolating high-purity RNA from in vitro cultured hybrid thermoplastic cell-laden hydrogel constructs is described.
MATERIALS
- RNAZol® RT reagent
- Ethanol, absolute (molecular biology grade)
- Isopropanol, absolute (molecular biology grade)
- 75% (v/v) ethanol (molecular biology grade)
- Ultrapure water (ddH2O) (molecular biology grade)
- Tris-buffered saline, pH 7.4 (TBS) (molecular biology grade)
- 0.5 mL microcentrifuge tubes (RNAse free)
- 1.5 mL microcentrifuge tubes (RNAse free)
- 2 mL microcentrifuge tubes (RNAse free)
- 2 mL microcentrifuge tubes with corundum beads (1 – 3 mm)
- Disposable pellet pestles (RNAse free)
- 0.1 – 10 µL filtered micropipette tips (RNAse free)
- 5 – 200 µL filtered micropipette tips (RNAse free)
- 100 – 1000 µL filtered micropipette tips (RNAse free)
- 100 mm diameter petri dishes (sterile)
- Scalpel blades (sterile)
EQUIPMENT
- Vortex
- Micropipettors
- Tweezers (sterile)
- Scalpel handle (sterile)
- Chemical fume hood
- Microtube centrifuge (12.000 xg)
- UV-Visible microvolume spectrophotometer
METHODS
Follow institutional and local guidelines for the safe handling and disposal of biological agents and chemical reagents. Unless otherwise stated, use RNAse free plasticware during all steps of the procedure.
- Prepare 75% (v/v) ethanol from absolute ethanol and ddH2O.
- Remove the tissue engineered scaffold from the cell culture medium, transfer to a sterile 100 mm Petri dish, and dissect into 1 mm3 pieces using a sterile scalpel.
- Transfer the dissected sample with the help of sterile tweezers into a 2 mL microcentrifuge tube containing RNAZol® RT reagent. Use 1 mL of RNAZol® RT reagent per 200 µL of hydrogel.
- Add the whole content of a corundum bead tube to the 2 mL microcentrifuge tube containing RNAZol® RT reagent and the sample.
- Disrupt and homogeneize the sample with the help of a disposable pellet pestle and vortex. Intersperse 3 pestle disruptions with 3 x 2 minutes full-speed vortex homogeneizations.
- After disruption and homogeneization, samples can be stored at -20 ºC for up to one year. If samples are stored at -20 ºC, thaw and equilibrate them at room temperature before continuing to the following step.
- Add 200 µL of ddH2O per each mililiter of RNAZol® RT reagent employed for sample preparation. This will make up a total volume of 1.4 mL (1 mL RNAZol® RT reagent + 200 µL hydrogel + 200 µL ddH2O).
- Shake vigorously for 15 seconds and allow to stand at room temperature for 15 minutes.
- Centrifuge at 12.000 xg for 15 minutes at room temperature. Centrifugation separates the mixture into a pellet containing DNA, proteins, polysaccharides and solids; and an upper supernatant containing RNA.
- Transfer supernatant (750 µL per 1.4 total volume homogenate) to a 1.5 mL microcentrifuge tube, leaving a layer of the supernatant above the DNA/protein pellet.
- Add an equal volume of isopropanol to the recovered supernatant in order to precipitate RNA. Allow to stand for 10 minutes at room temperature.
- Centrifuge at 12.000 xg for 10 minutes at room temperature. The RNA will precipitate as a white pellet on the bottom of the tube.
- Wash RNA pellets with 0.6 mL 75% (v/v) ethanol thrice. Vortex for a few seconds and centrifuge at 8.000 xg for 3 minutes at room temperature for every ethanol wash.
- Remove alcohol solution completely with a micropipette, and air dry any remain of ethanol solution from the tubes to avoid interferences with downstream applications.
- Solubilize the RNA pellet in a volume of 30 – 50 µL ddH2O. Vigorous pipetting may be neccesary in order to achieve complete RNA solubilization. After solubilisation, keep samples on ice.
- Dilute a small aliquot of the sample 1/2 with TBS for UV-visible spectrophotometric quantification. Prepare a blank sample by diluting TBS 1/2 with ddH2O.
- Measure absorbance of the prepared aliquots at 230, 260 and 280 nm. If possible, perform a spectral scan from 220 to 350 nm with baseline correction at 340 nm. Calculate the absorbance ratios at 260/280 nm, and 260/230 nm for estimating purity.
- For long-term storage, samples should be kept at -80 ºC.
RESULTS
Following the abovementioned protocol, a final preparation of RNA is obtained, free of DNA and proteins. Pure RNA preparations should should have a 260/280 nm ratio of 1.7- 2.1 and a 260/230 ratio of 1.6 to 2.3.
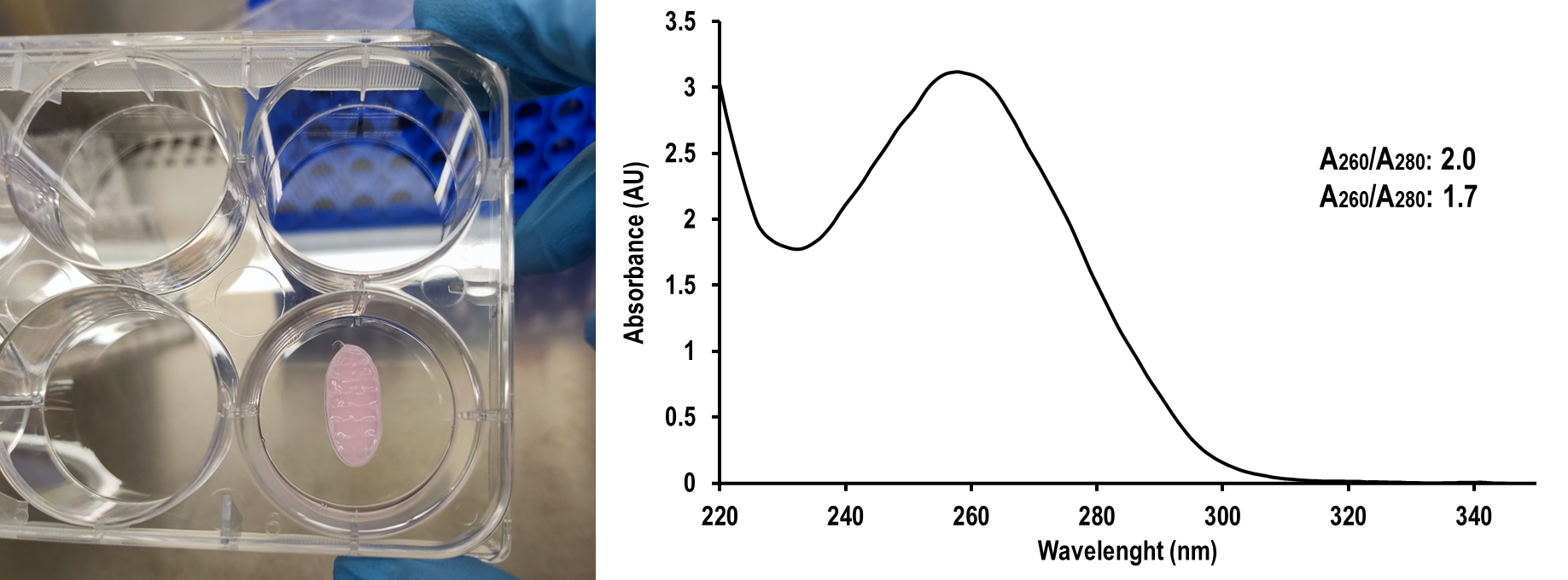
Figure 1. In vitro cultured hybrid thermoplastic cell-laden hydrogel tissue-engineered constructs (left). UV-visible absorbance spectrum of RNA isolated following the abovementioned protocol (right).
| Number | Category | Product | Amount |
|---|
